In GLM-GWAS calculations, the imputation step is optional, not mandatory. In this regard, the GLM-GWAS calculation procedure provides the following two filtering options: (1) Missing ID %: Removes samples with a missing rate higher than the set value (0-1).(2) Missing SNP %: Removes SNP markers with a missing rate higher than the set value (0-1).
Meanwhile, in LMM-GWAS calculations, both of the above two filtering options are performed during the imputation process.
Figure 1 shows the GLM-GWAS tab page when it’s selected. When you choose a phenotype file written in CSV format, the header values of that phenotype file will be listed in the “Columns” text box.
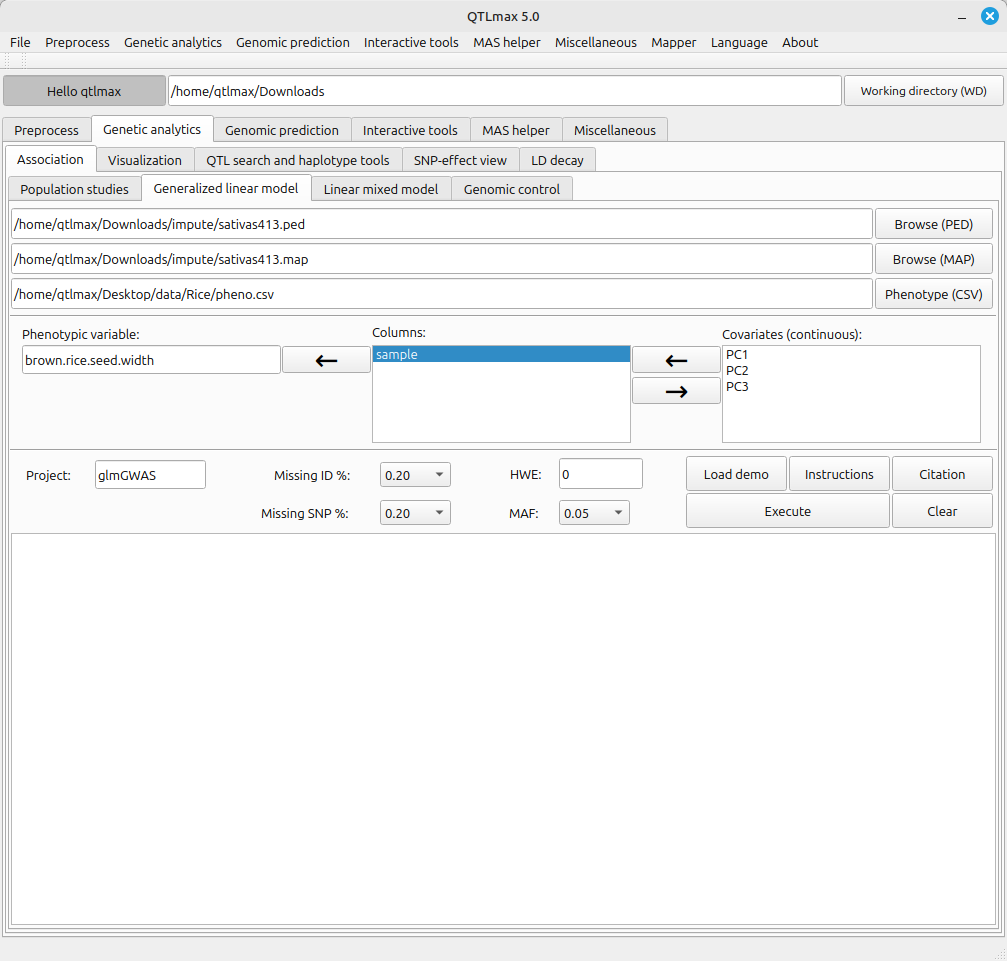
(Figure 1)
In addition to the filtering options mentioned above, the GLM-GWAS tab page requires a few other settings, and each configuration method is as follows:
- Send the header value that specifies the sample to the “Sample column” text box.
- Send the header value that specifies the phenotype value to the “Phenotype column” text box.
- Send the header values that specify the PCA values to the “Covariate (continuous)” text box. (e.g., PC1, PC2, PC3)
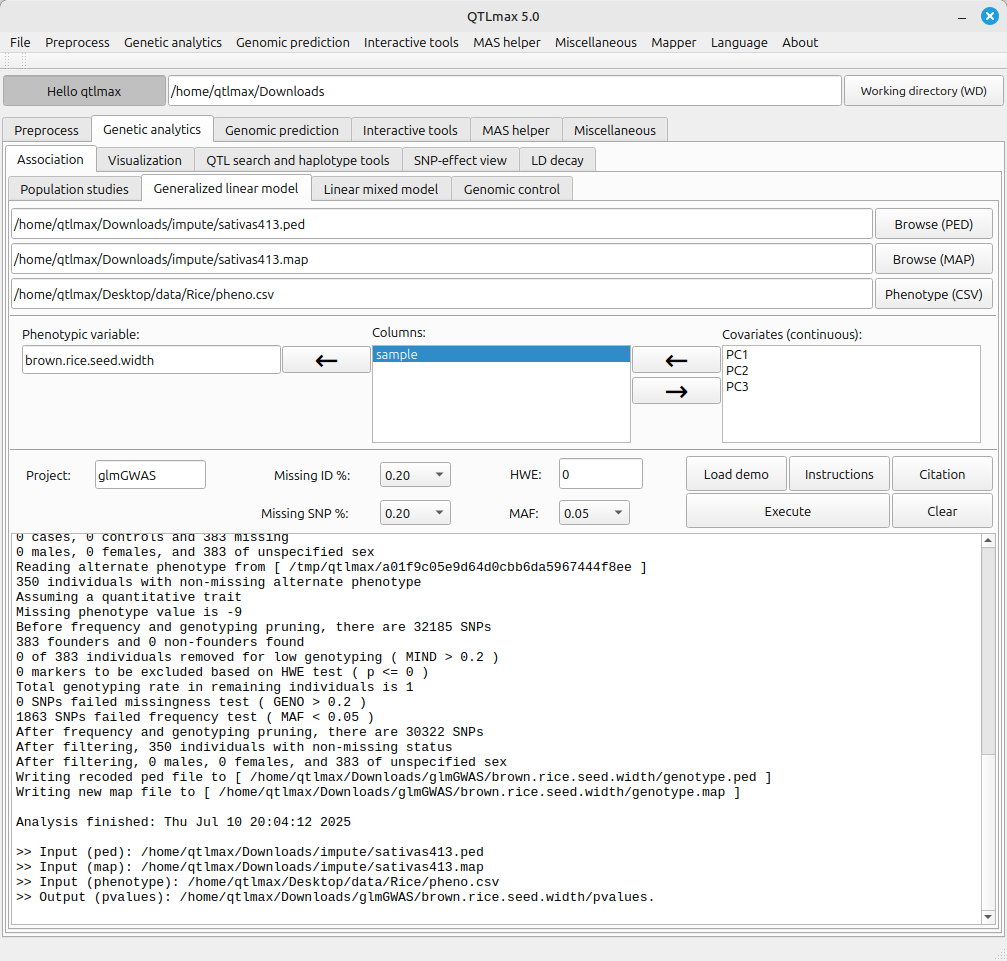
(Figure 2)
The path to the GLM-GWAS calculation result file is as follows:
Working directory > Project > Trait folder