Figure 1 shows the tab page related to backcrossing. The input data for backcross selection is the “similarity_matrix.csv” file obtained using QTLmax.
- To view a detailed explanation on how to generate the “similarity_matrix.csv” file, please click the following link: https://open.qtlmax.com/guide/index.php/2025/07/03/genetic-similarity-matrix-computation/
When you select the “similarity_matrix.csv” file, the names of the individuals included in that file will be displayed as a list (Figure 1).
(Figure 1)
If you double-click the recurrent parent, its name will automatically appear in the text box. Enter a threshold value in the “Equal to or greater than” text box and click the [Execute] button to display a list of individuals that satisfy genetic similarity with the recurrent parent at or above the threshold value. (Figure 2)
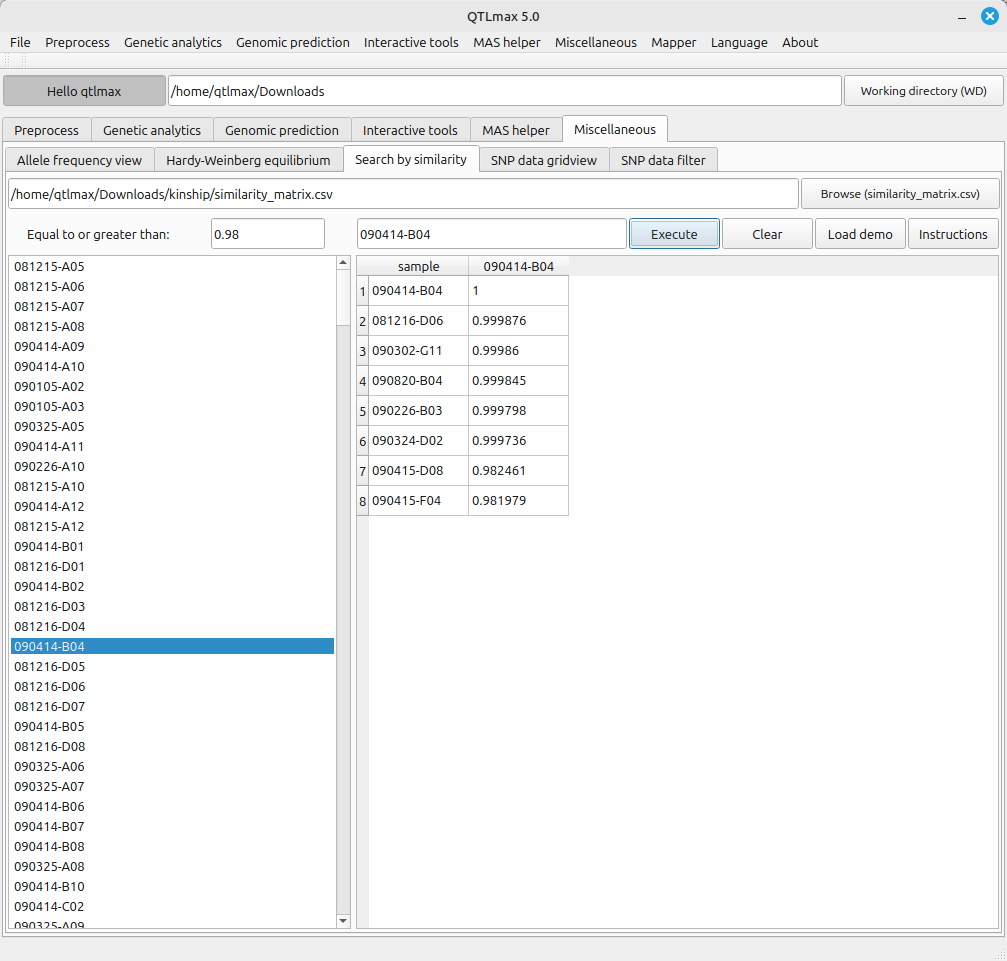
(Figure 2)