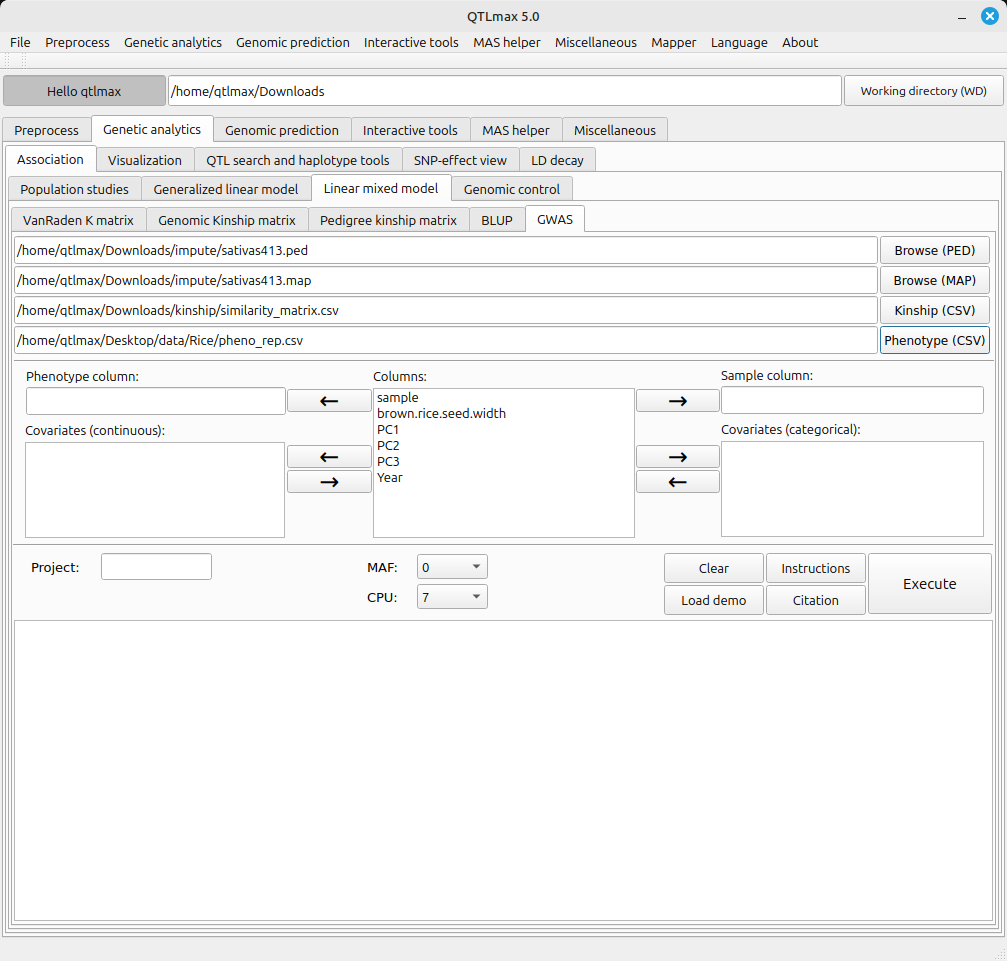
Figure 1 shows the LMM-GWAS tab selected. You need to select files a pair of imputed PED and MAP files, a matrix file for kinship matrix (e.g., genetic similarity matrix, pedigree kinship matrix, genomic kinship matrix) and a CSV file containing a phenotype file merged with a PCA/ADMIXTURE output file; when you choose this file in CSV format, its header values will be listed in the “Columns” text box. (Figure 1)
(Figure 1)
The LMM-GWAS tab requires a few settings. Here’s how to configure each one:
- Send the header value that specifies the sample to the “Sample column” text box.
- Send the header value that specifies the phenotype value to the “Phenotype column” text box.
- Send the header values that specify the PCA values (e.g., PC1, PC2, PC3) to the “Covariate (continuous)” text box.
- If there are any categorical header values that specify environmental factors (e.g., location, year, replication, treatments), send them to the “Covariates (categorical)” text box.
In the CPU field of dropdown menu, you can select the number of CPUs to use for parallel computation. For example, setting the CPU value to 10 means you will use 7 CPUs for parallel calculations. Naturally, the more CPUs you use, the faster the computation speed will be.
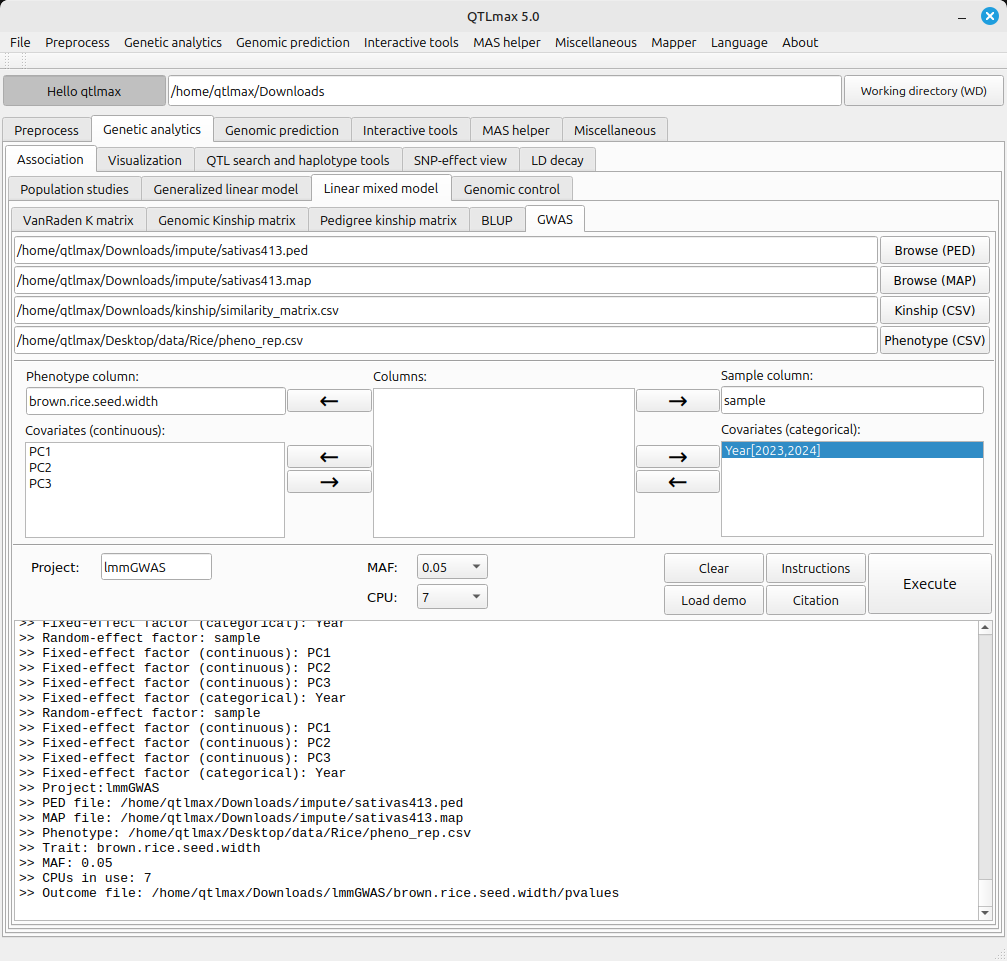
(Figure 2)
The path to the LMM-GWAS calculation result files is as follows:
[Working directory] > [Project folder] > [trait folder]
One thought on “GWAS procedure (linear mixed model)”