Figure 1 shows the tab page for the imputation job. For imputation, you need a file in VCF format or VCF.GZ format (a compressed VCF file). The following settings are required for the imputation job:
(1) Missing ID %: Removes samples with a missing rate higher than the set value (0-1).
(2) Missing SNP %: Removes SNP markers with a missing rate higher than the set value (0-1).
(3) HWE: This refers to the Threshold value for Hardy-Weinberg equilibrium filtering. If there are many zeros after the decimal point, you can use “e” for notation. For example, a value of 0.0000000001 can be expressed as 1e-10.
(4) MAF: Minor allele frequency.
(5) Burn-in
(6) Iterations
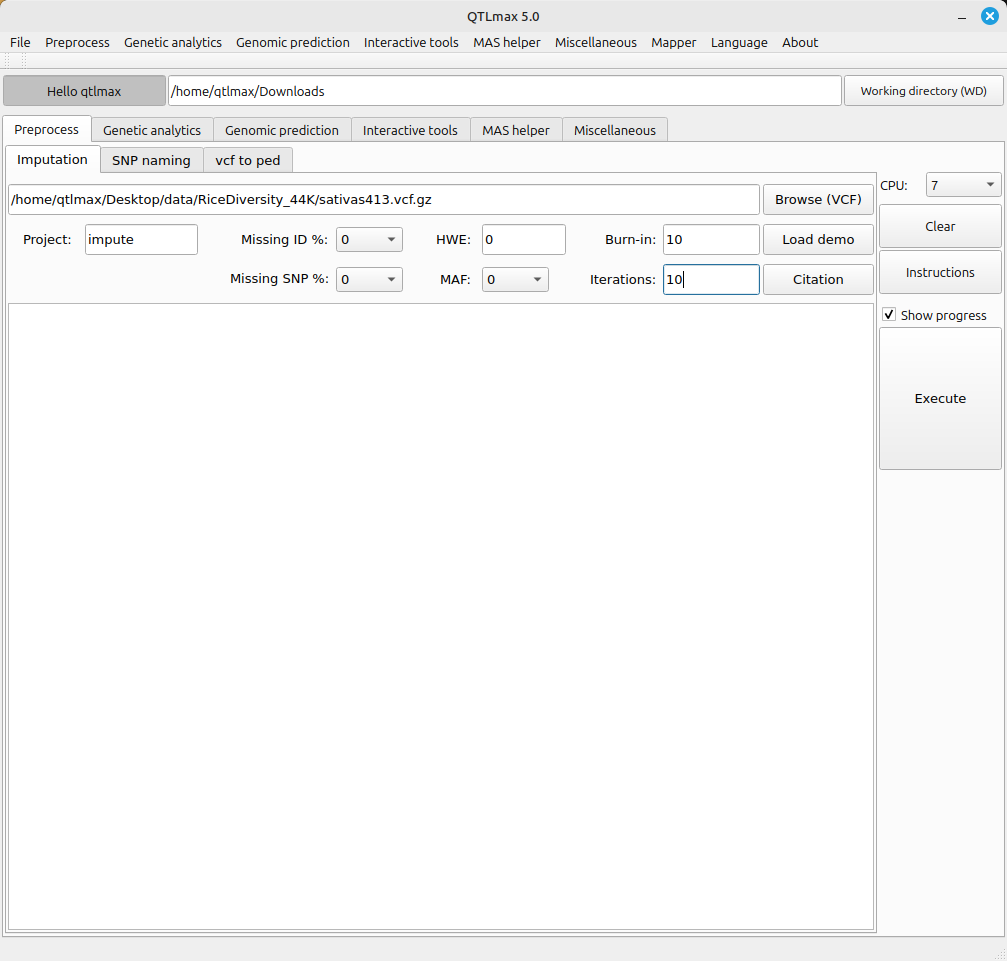
(Figure 1)
After entering all the settings and checking the “Show progress” box, clicking the [Execute] button will bring up a pop-up terminal, displaying the imputation process as it is being performed (Figure 2).
(Figure 2)
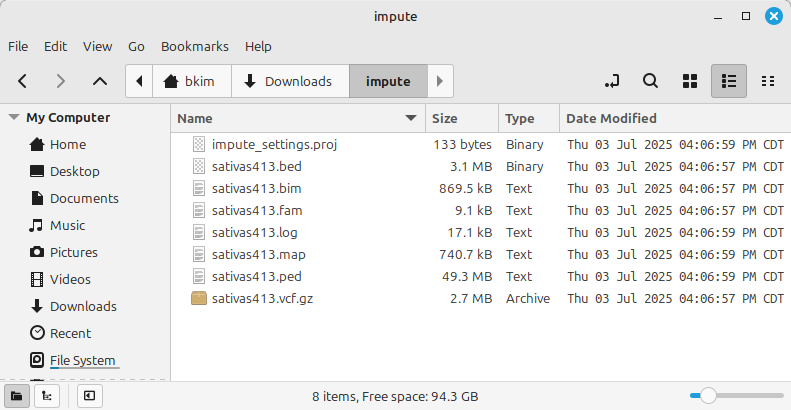
(Figure 3)
Shown in Figure 3 is the imputation result files, which are of three types:
- PLINK format (*.PED, *.MAP)
- PLINK binary format (*.bed, *.bim, *.fam)
- Compressed VCF (*.vcf.gz)
Among the three genomic data formats above, the format required by QTLmax is #1. All three file formats contain the same information and can be interconverted.
One thought on “Imputation”