This post explains how to predict subpopulation class for individuals given known genomic analytics, using deep learning algorithms.
Figure 1 shows the “Deep-learning classification” tab selected in QTLmax Super.
(Figure 1)
The data format for input into the “Deep-learning classification” calculation is as shown in Figure 2. As you can see, the input file has a “Subpopulation” column added to the right side of the PCA result file. You’ll notice that while subpopulation information is not present for the individuals at the top of the file, it is displayed for the individuals at the bottom.
The deep learning algorithm will first perform training on the individuals for whom subpopulation information is available, and then proceed to predict subpopulation values for the individuals without that information.
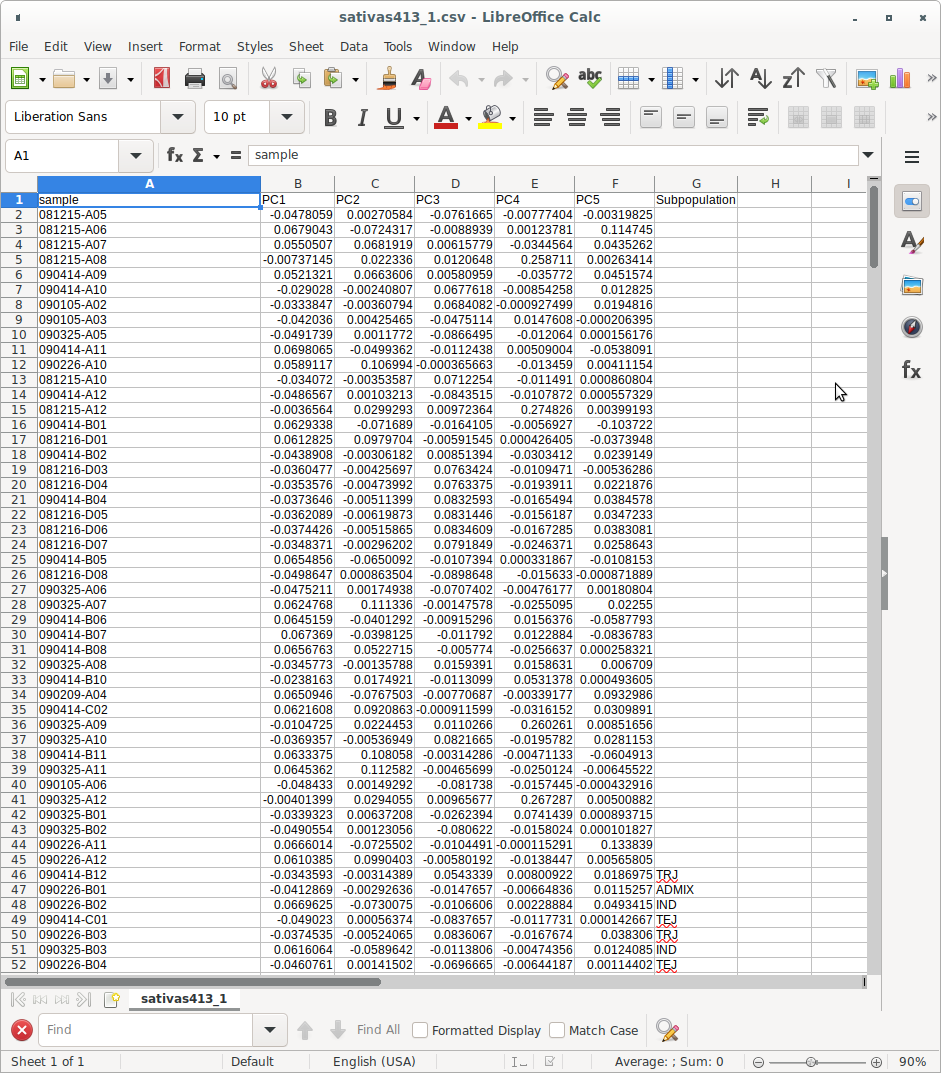
(Figure 2)
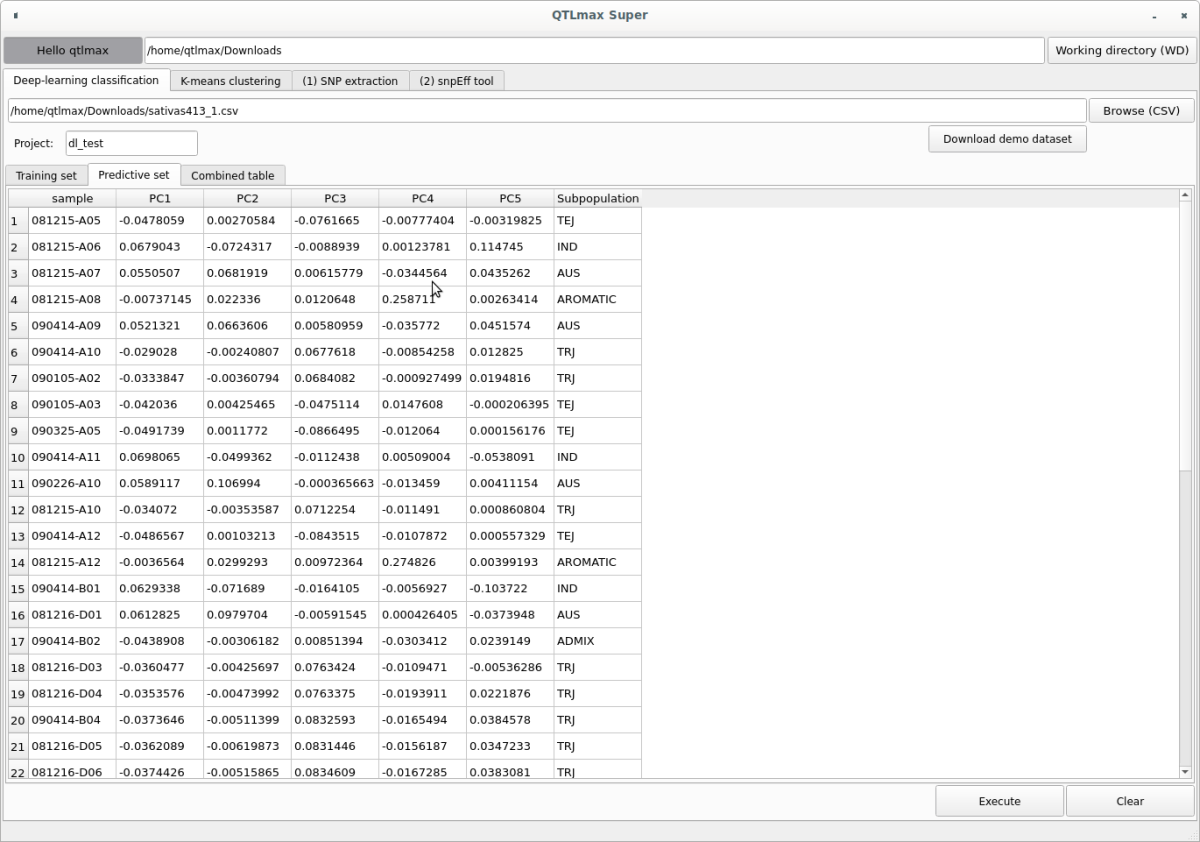
When you enter the project name and select the input file, the “Training set” tab will display individuals with subpopulation information, and the “Predictive set” tab will show individuals without subpopulation information (Figures 3, 4).
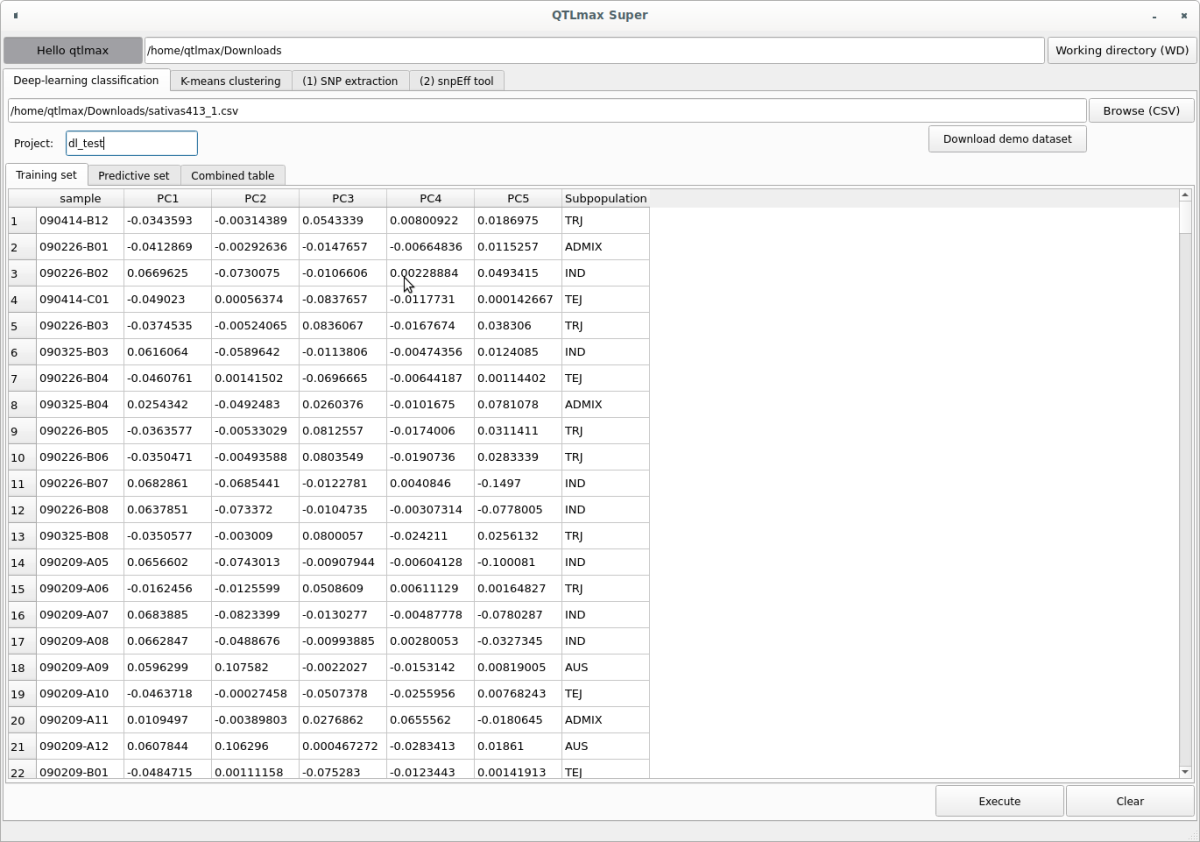
(Figure 3)
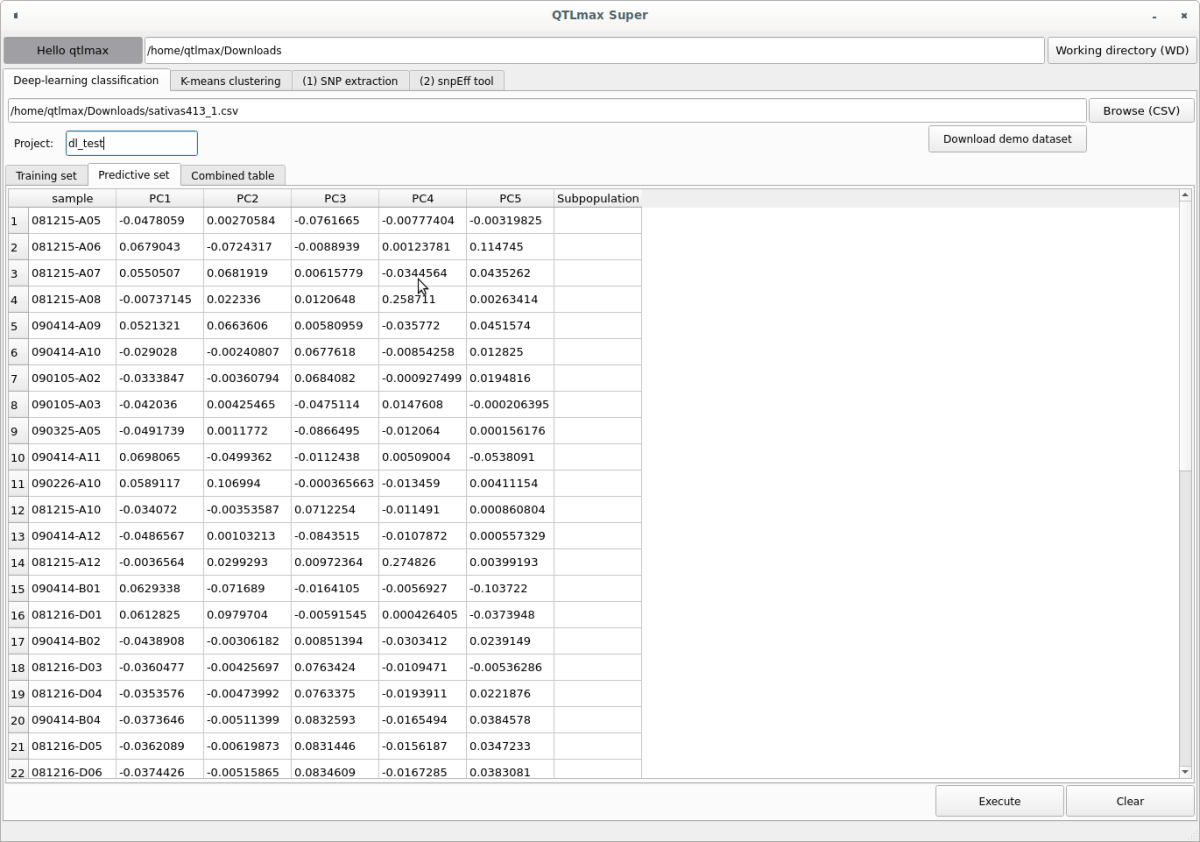
(Figure 4)
When you click the [Execute] button, the deep learning task runs, and you’ll see the subpopulation information generated and displayed for the individuals in the “Predictive set” (Figure 5).
The “Combined table” tab simply shows a table that merges the tables from the “Training set” tab and the “Predictive set” tab.
(Figure 5)